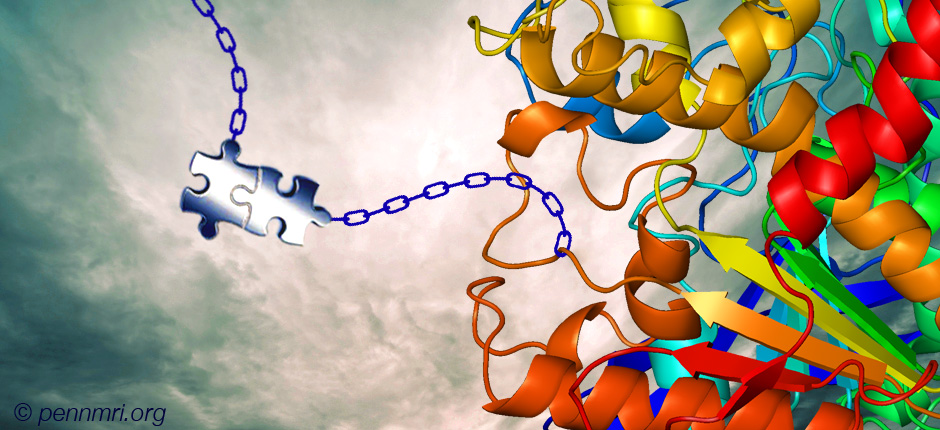
A wide variety of functional group handles have been incorporated into proteins, however only a few have the potential of being engaged in ligation reactions in vivo. Of particular challenge in this setting are the high dilution reaction conditions requiring relatively high ligation reaction rates. Two recent reports successfully employ trans-cyclooctene (TCO) modified proteins to achieve site-specific protein modifications in living cells.
A wide variety of functional group handles have been incorporated into proteins, however only a few have the potential of being engaged in ligation reactions in vivo. Of particular challenge in this setting are the high dilution reaction conditions requiring relatively high ligation reaction rates that must be accompanied by low side reaction rates in the context of the wide range of functional groups present in living systems. In addition to being relatively resistant to attrition due to metabolism, ligation reagents must have favorable absorption, distribution, and excretion profiles, collectively their pharmacokinetic properties.
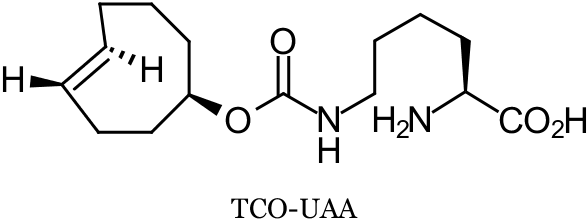
Two recent reports successfully employ trans-cyclooctene (TCO) modified proteins to achieve site-specific protein modifications in living cells. In a collaboration between Lemke, Schultz, and co-workers at the European Molecular Biology Laboratory (EMBL, Heidelberg, Germany) and Wieβler at the German Cancer Research Center (DKFZ, Heidelberg, Germany), an unnatural amino acid (UAA) TCO-derivative (TCO-UAA, see below) was genetically encoded by an engineered tRNA/synthetase pair through the suppression of the Amber stop codon. This was accomplished in both E. coli and mammalian cells. TCO-tagged proteins were labeled with fluorescent tetrazines (Tz) via the strain-promoted inverse electron-demand Diels-Alder cycloaddition (SPIEDAC) TCO-Tz ligation by exposing the living cells to Tz-dyes. In E. coli, TCO-UAA tagged green fluoresecent protein (GFP) was labeled with a tetramethylrhodamine-Tz conjugate (TAMRA-Tz) with an exceptionally high rate constant of 35000 M-1s-1. In mammalian HeLa cells, while the rate constant was not determined, the labeling of TCO-UAA tagged maltose binding protein (MBP) with the pentamethine cyanine dye Cy5-Tz could be recorded after only 5 minutes. In addition to these UAA expression and labeling experiments, the chemical orthogonality of the TCO-Tz ligation (SPIEDAC) to the strain-promoted alkyne-azide cycloaddition (SPAAC) was demonstrated by the double labeling of a mixture of two E. coli cultures expressing either TCO-UAA or a cyclooctyne-UAA. This mixture was
treated first with TAMRA-azide engaging only the cyclooctyne-tag (via SPAAC), followed by treatment with coumarin-Tz to label the TCO-tag (via SPIEDAC). Selective labeling of each of the tags was qualitatively confirmed by the presence of a cell population with an even distribution of only two cell types with either coumarin or TAMRA-specific fluorescence.
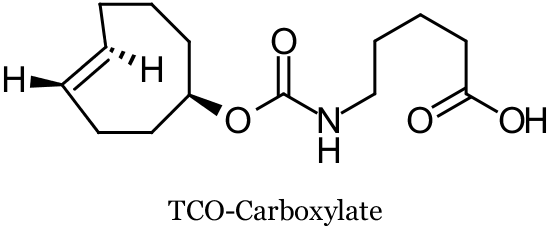
In a collaboration between Ting and co-workers at Massachusetts Institute of Technology and Fox and co-workers at the University of Delaware, TCO-modified proteins were prepared using probe incorporation mediated by enzymes (PRIME), a post-translational protein labeling method. These TCO-tags were fluorescently labeled both on cell surfaces and inside living cells by their chemoselective ligation with Tz-fluorophore conjugates (via SPIEDAC). Specifically, a mutant of the enzyme lipoic acid ligase (LplA) was used to catalyze the coupling of TCO-carboxylate derivatives (for example TCO-Carboxylate, see below) to recombinant proteins fused to a 13-amino acid LplA acceptor peptide. TCO-Carboxylate was thus incorporated into LAP-tagged low-density lipoprotein receptor expressed on the cell surface of human embryonic kidney 293T cells (HEK cells), LAP-tagged neuroligin-1 on the surface of rat neurons, LAP tagged nuclear-localized blue fluorescent protein (nuclear LAP-BFP) inside HEK cells, and LAP-tagged β-actin and vimentin filaments in COS-7 cells. Several fluorophore tetrazine conjugates were designed and tested varying the tetrazine moiety to optimize for SPIEDAC reactivity and chemical stability, and included the fluorophores tetramethylrhodamine (TAMRA), Alexa Fluor 647, fluorescein, and cell-permeable fluorescein diacetate. On the cell surface, fluorogenic labeling was achieved in 3-5 minutes. Intracellular labeling was hampered by Tz-probe cross reactivity, requiring off-target dye washout.
For more information about these reagents and techniques, please see the references below. For related topics please explore our collection of articles.
References:
- “Amino Acids for Diels-Alder Reactions in Living Cells” Plass, T.; Milles, S.; Koehler, C.; Szymański, J.; Mueller, R.; Wieβler, M.; Schultz, C.; Lemke, E. A. Angew. Chem. Int. Ed. 2012, 51, 4166-70.
- “A Need for Speed: Genetic Encoding of Rapid Cycloaddition Chemistries for Protein Labeling in Living Cells” Schmidt, M. J.; Summerer, D. ChemBioChem 2012, 13, 1553-7.
- “Genetically Encoded Tetrazine Amino Acid Directs Rapid Site-Specific In Vivo Bioorthogonal Ligation with trans-Cyclooctenes” Seitchik, J. L.; Peeler, J. C.; Taylor, M. T.; Blackman, M. L.; Rhoads, T. W.; Cooley, R. B.; Refakis, C.; Fox, J. M.; Mehl, R. A. J. Am. Chem. Soc. 2012, 134, 2898-901.
- “Diels-Alder Cycloaddition for Fluorophore Targeting to Specific Proteins inside Living Cells” Liu, D. S.; Tangpeerachaikul, A.; Selvaraj, R.; Taylor, M. T.; Fox, J. M.; Ting, A. Y. J. Am. Chem. Soc. 2012, 134, 792-5.
- “Design and Synthesis of Highly Reactive Dienophiles for the Tetrazine-trans-Cyclooctene Ligation” Taylor, M. T.; Blackman, M. L.; Dmitrenko, O.; Fox, J. M. J. Am. Chem. Soc. 2011, 133, 9646-9.
- “(E,E)-1,5-Cyclooctadiene: a small and fast click-chemistry multitalent” Stöckmann, H.; Neves, A. A.; Day, H. A.; Stairs, S.; Brindle, K. M.; Leeper, F. J. Chem. Comm. 2011, 47, 7203-5.
- “Tetrazine Ligation: Fast Bioconjugation Based on Inverse-Electron-Demand Diels-Alder Reactivity” Blackman, M. L.; Royzen, M.; Fox, J. M. J. Am. Chem. Soc. 2008, 130, 13518-9.